Nannochloropsis genomes, what have we learnt?
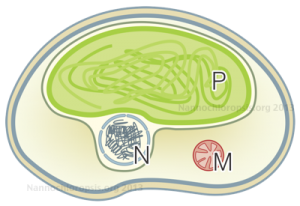
Schematic representation of Nannochloropsis cell by Andrea Telatin
Thanks to the sequencing of the genomes of two strains of Nannochloropsis gaditana [1][2] and two strains of Nannochloropsis oceanica [3][4] we now have the reference sequences to design molecular biology experiments on this microorganism. Moreover we have learnt a great deal of information, opened up new interrogatives .. and more will come analysing the data available though this portal and through the other web resources: N. gaditana CCMP526 genome and N. oceanica CCMP1779 genome.
Nannochloropsis genome is small, about 28-29 Mega bases, and very compact: there is on average 1 gene each 2.7 kilo bases in N. gaditana B-31, the genes are long on average 1.2 kilo bases and they contain very few introns.
The proteins predicted in the two species are in large part similar, about 80% of the proteins of each species are found in clusters of homologous proteins, ~60% of which accomodate proteins common to the two species. Further studies will help ruling out the limits and the inaccuracies of the gene predictions allowing to focus on the actual differences between the two species.
The pathways leading to the synthesis of cellulose and sulfated fucans and to the remodelling of cellulose in the cell wall have been identified both in N. gaditana and in N. oceanica, casting a light on the molecular composition of the cell wall and suggesting possible targets of genetic modification.
Thank you!
We currently have more than 1.000 unique visitors per month, accounting for roughly 18,000 pageviews per month. Visitors are spread across the four continents… Thank you for keeping alive Nannochloropsis.org. We are doing our best to update this site and we are also preparing new resources (to be released soon).

This week’s visits
Recent Comments